Welcome to GenePalette!
GenePalette is a powerful cross-platform and cross-species desktop application for genome sequence visualization and navigation. Users can download segments of genome sequence from NCBI’s GenBank database from a variety of organisms (e.g. yeast, human, fly, worm, mouse, plants), preserving the gene annotation that is associated with that sequence. Sequence elements of interest (transcription factor binding sites, other regulatory motifs, restriction enzyme sites, primer sequences, SNPs, microsatellites, etc.) can be searched for and identified in the loaded sequence, and then clearly visualized within a colorful graphical representation of gene organization and intron/exon structure. Sequence alignment functions facilitate phylogenetic footprinting and cross-species comparative studies.
GenePalette in the literature
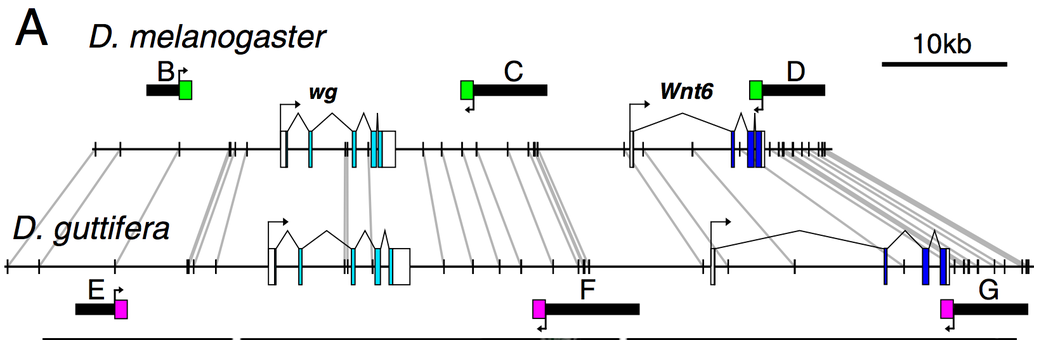
Figure from Koshikawa et al., PNAS 2015 Depicting the relationship between the wingless locus of Drosophila melanogaster and Drosophila guttifera
Figure from Koshikawa et al., PNAS 2015 Depicting the relationship between the wingless locus of Drosophila melanogaster and Drosophila guttifera
News
July, 2017 - Publication describing new capabilities, and new version, 2.1.2
We are proud to announce a new publication that describes the new version of GenePalette in Developmental Biology! The paper can be found here. If you use GenePalette in your work, please cite this paper
We have also released a new version (2.1.2) that fixes a minor bug with OligoList features
We have also released a new version (2.1.2) that fixes a minor bug with OligoList features
May, 2017 - GenePalette version 2.1
We are pleased to announce a new version of the software that implements a new module which automates the acquisition of orthologous sequences from the UCSC genome database. Click here to see a video demonstration of the new module in action! In addition, we have made the following improvements:
- Fixed bugs associated with the sequence comparison functions
- Updated GenBank access protocols to accommodate their recent shift to https protocols.
- We have updated the downloadable genome sequences available, and now include recent mouse, human, fly, nematode, and Arabidosis genome builds
- Fixed bugs associated with the sequence comparison functions
- Updated GenBank access protocols to accommodate their recent shift to https protocols.
- We have updated the downloadable genome sequences available, and now include recent mouse, human, fly, nematode, and Arabidosis genome builds
This work is generously supported by the NIH (R35GM14196) and a grant from the NSF (2129903)